![PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/af80a5d2beee749cff2f5d98dc4a8f0dfdd48074/4-Table3-1.png)
PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar

Photometric Quantification of Proteins in Aqueous Solutions via UV-Vis Spectroscopy - Eppendorf Handling Solutions

Molecules | Free Full-Text | Evaluation of Peptide/Protein Self-Assembly and Aggregation by Spectroscopic Methods

Observed and predicted molar absorption coefficients at 280 nm for 80... | Download Scientific Diagram

Prediction of Molar Extinction Coefficients of Proteins and Peptides Using UV Absorption of the Constituent Amino Acids at 214 nm To Enable Quantitative Reverse Phase High-Performance Liquid Chromatography−Mass Spectrometry Analysis | Journal

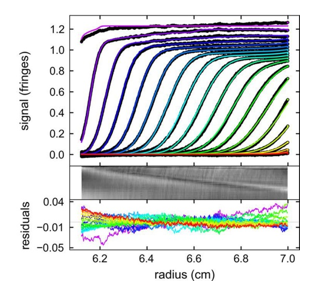
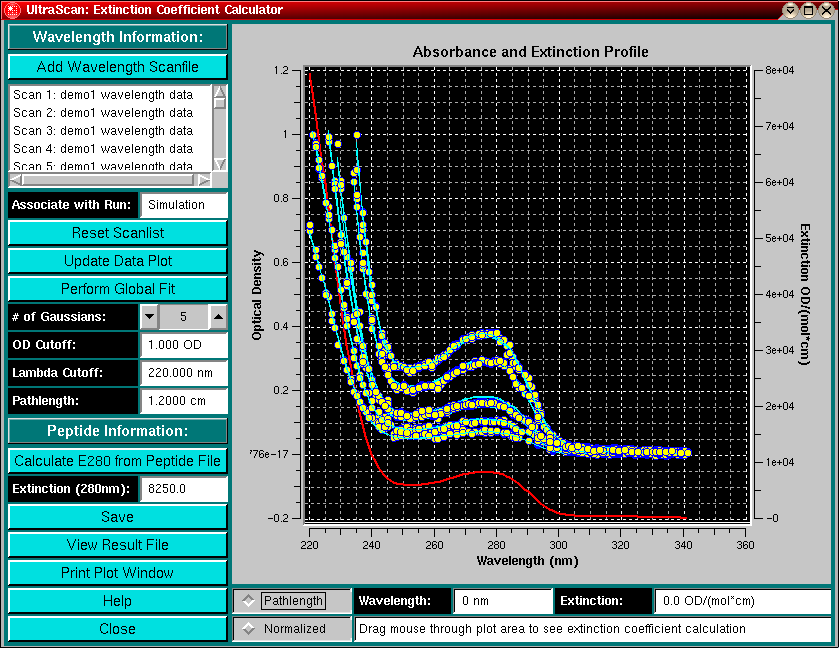
Precise determination of protein extinction coefficients under native and denaturing conditions using SV-AUC | SpringerLink

Prediction of Molar Extinction Coefficients of Proteins and Peptides Using UV Absorption of the Constituent Amino Acids at 214 nm To Enable Quantitative Reverse Phase High-Performance Liquid Chromatography−Mass Spectrometry Analysis | Journal
Engrailed and Hox homeodomain proteins contain a related Pbx interaction motif that recognizes a common structure present in Pbx. - Abstract - Europe PMC
![PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/af80a5d2beee749cff2f5d98dc4a8f0dfdd48074/2-Table1-1.png)
PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar
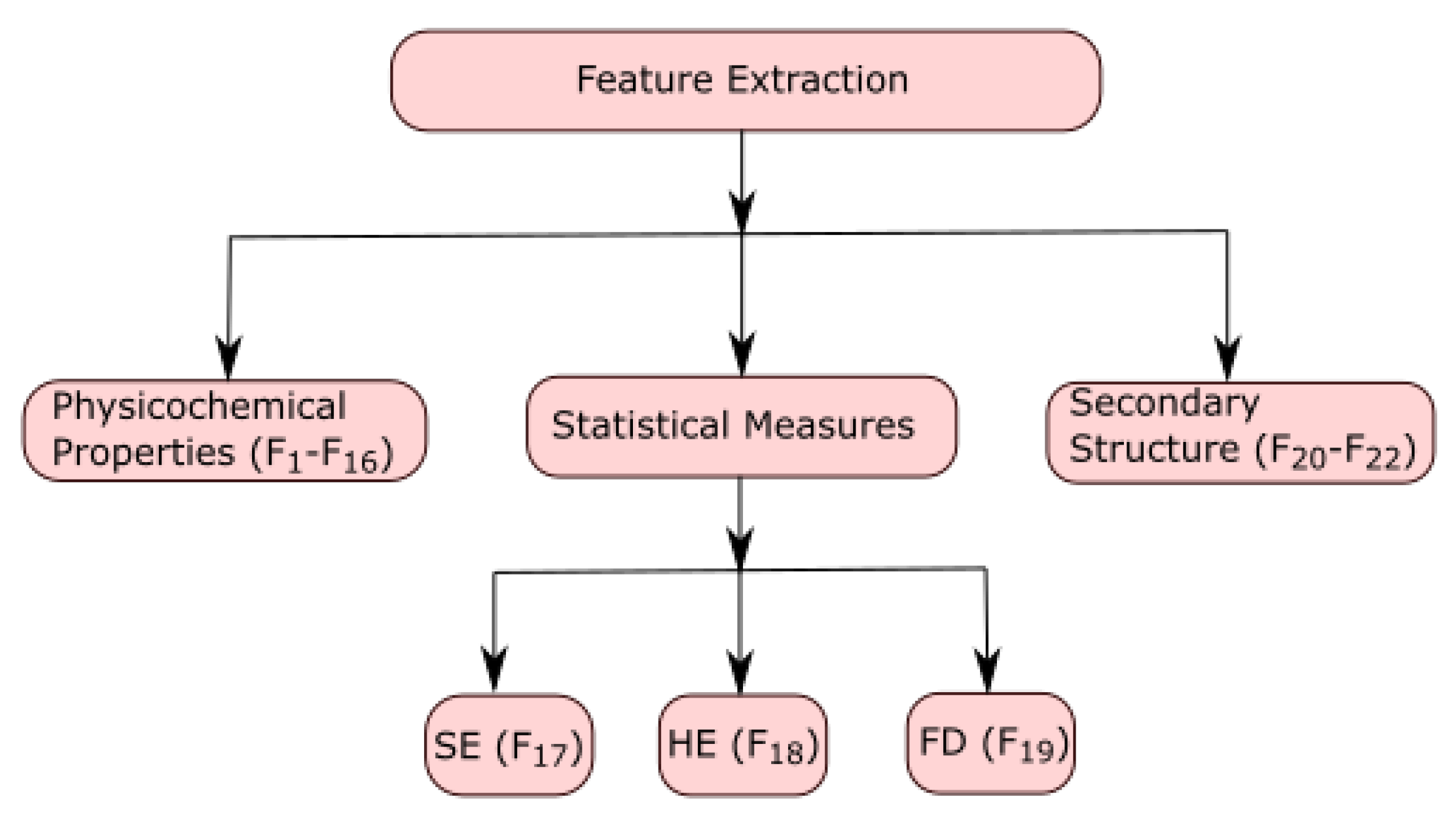
Mathematics | Free Full-Text | Unsupervised Learning for Feature Representation Using Spatial Distribution of Amino Acids in Aldehyde Dehydrogenase (ALDH2) Protein Sequences